Mapping the genetic architecture of severe male infertility
Male infertility due to spermatogenic failure is a common disorder found in 1% of men. Although the contribution of genetic predisposition is considered to be substantial, the genetic causes of severe male infertility have largely remained elusive. It is feasible that rare patient-specific mutations across a multitude of genes essential for germ cell development may lead to the manifestation of the disease.
GEMINI, a multi-center study comprised of andrology investigators from 20 centers in nine countries, has recruited a cohort of 1850 patients and over 2000 controls to date (Spring 2024). It is currently funded by two major NIH grants (R01 HD078641 and P50 HD096723-01) and is in its ninth year of funding.
Current ongoing research activities are predominantly focused on identifying and describing the genetic composition of non-obstructive azoospermia (NOA) as the most severe type of male infertility. Whole-exome sequencing approach is applied to identify rare genetic alterations among the cases followed by variant prioritization and functional validation of most relevant findings. GEMINI has lead or contributed to the discovery of over 30 genes associated with male infertility over the past 6 years, reported in over 15 peer-reviewed publications.
The long-term goals of GEMINI project include expanding the network and research interest to other types of male but also female infertility phenotypes.
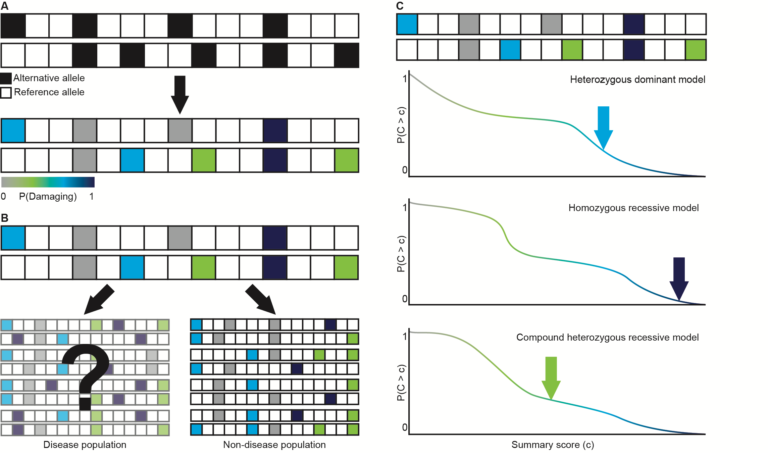
Variant prioritization tool enabling a case-by-case analysis without a need for matching control samples. Wilfeft et al. 2016, Nat Genet
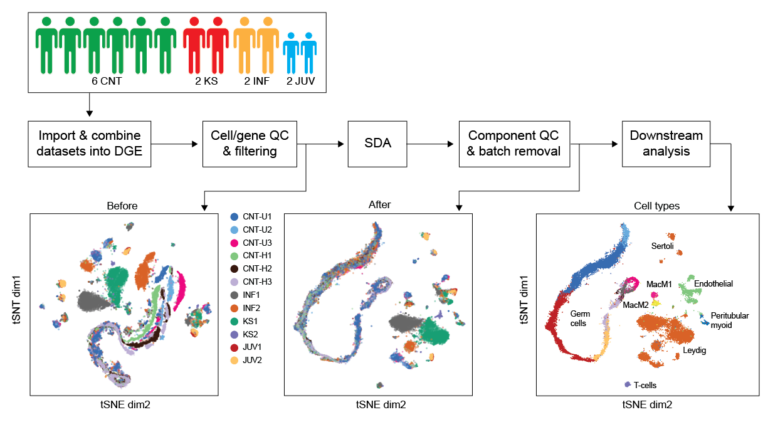
Genomic analysis of patient biopsies using single-cell and spatially-resolved transcriptomics. Mahyari et al. 2021 AJHG
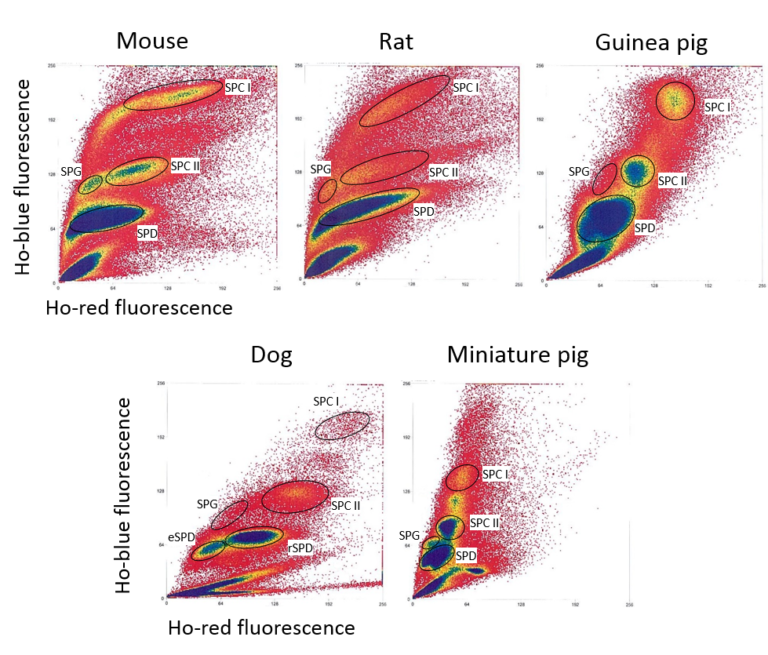
Flow cytometric sorting of five subpopulations of germ cells from various mammalian species in a manner compatible with downstream high-throughput studies. Lima et al. 2017, J Vis Exp
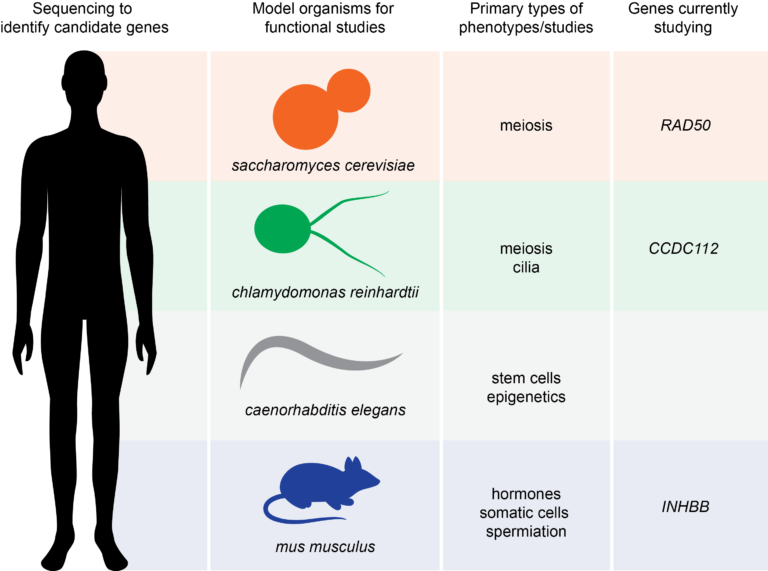
In-house and collaborative efforts for functional validation of human disease variants with appropriate methods (e.g. knock-out or CRISPR-Cas9) and model organisms (including mouse and zebrafish).